TargetDB
TargetDB is a tool to quickly query multiple publicly available databases and provide an integrated view of the information available about potential targets. A quick binding pocket search is also performed (using fpocket).
Tutorial: A document containing details on the methods used can be found HERE (pdf)
Installation
Virutal Environment Set-up
We recommend running the TargetDB scripts in a virtual environment to self-contain the application and to avoid interference with packages and libraries already installed on your system.
1) Dowload source code
git clone https://github.com/sdecesco/targetDB.git
2) Navigate to the targetDB directory and set up virtual environment
cd targetDB
virtualenv venv
3) Activate the virtual environment and install package dependencies
source venv/bin/activate
pip install -r requirements.txt
4) Run the druggability_report.py script to activate the GUI and use the TargetDB tool
python targetDB/druggability_report.py
Python compatibility
python version >= 3.4
Preferred python distribution : Anaconda 3
Database files installation
This package relies on the use of sqlite database to properly function.
Required database
- targetDB
You can download a copy of the old database HERE
A more recent version of the database has been created which can be downloaded HERE
We recommend using the most recent database for the most up-to-date results
This database contains all of the human genome genes that have a uniprot ID
System compatibility
- Linux
- Windows
- Mac (not tested)
Usage
targetDB package provides a user interface to use the tool: targetDB
targetDB
When using targetDB for the first time you will be asked to enter information about:
- targetDB sqlite database file
- path to save list output files
- path to save detailed target output files
- email address (used for pubmed searches if none provided no pubmed search will be run)
Those informations will be stored in ~/.targetdb/config.ini
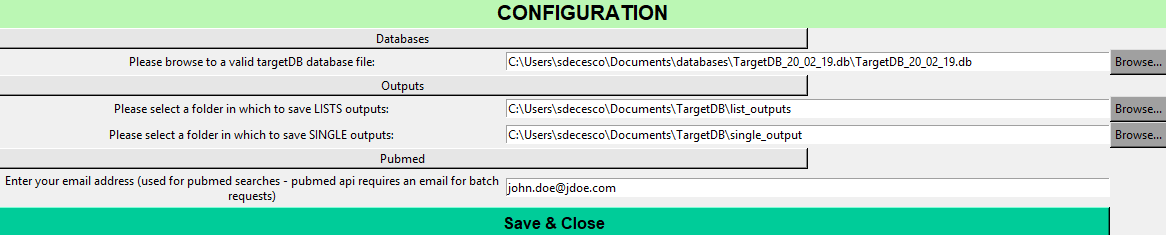
Once created it will automatically start the main user interface (as seen below)

Examples
TargetDB can be used in three modes:
- Single
- List
- Spider plot
Single mode outputs
A series of examples can be downloaded here (Excel file)
List mode outputs
In this example we have used a list of 95 targets provided by the AMP-AD consortium (LINK HERE)
You can download two version of the list with different weight used to construct the MPO Score
You can also find a list used to prioritize an entire class of proteins (Solute Carrier Protein)
A definition of all the columns in the output can also be downloaded
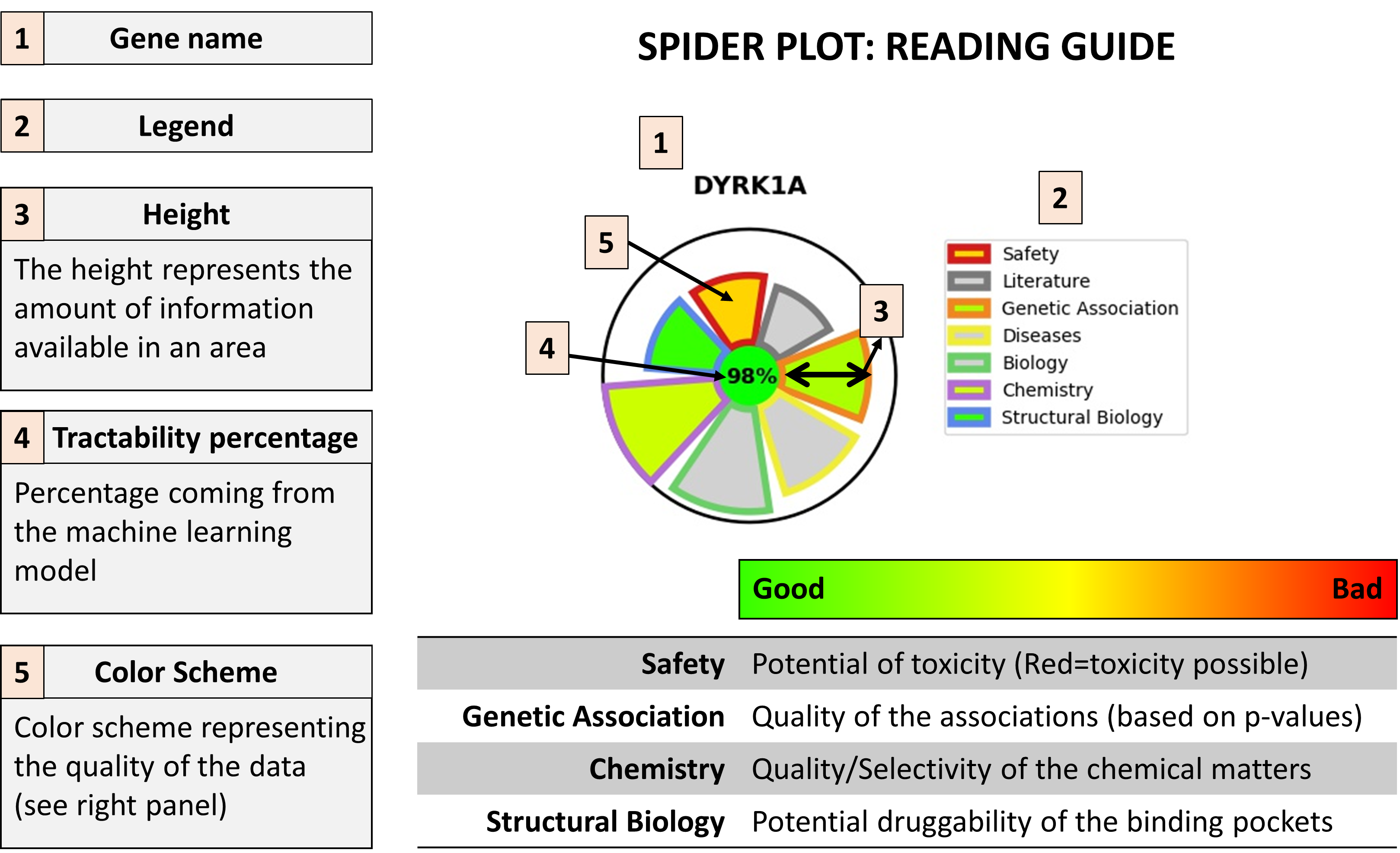
Spider Plots
A spider plot is here used to quickly give an idea of the area in which a target has strength and weaknesses

Here under a guide to help reading these plots:
Instructions to create a targetDB database from scratch
WARNING: This mode is not fully supported and we cannot guarantee it will execute without errors
System compatibility
- Linux
Database creation
target_DB
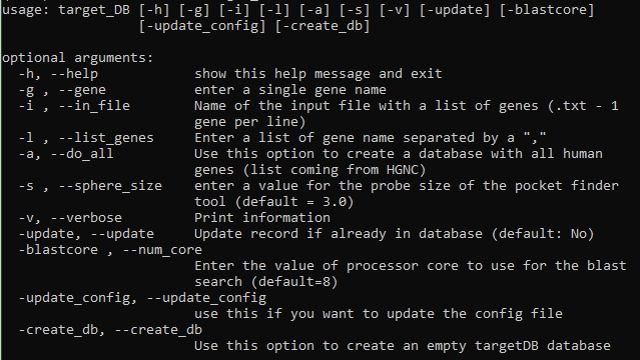
The list of required databases is :
- ChEMBL v25
ChEMBL sqlite database can be directly downloaded HERE
This mode will generate a targetDB database that can then be used in report mode
Other dependencies for the database creation mode
blast
This mode use blast locally to perform similarity search and sequence alignments
information to download and install blast locally can be found HERE
fpocket
In order to perform binding pocket searches and assess their druggability the program fpocket is used
Vincent Le Guilloux, Peter Schmidtke and Pierre Tuffery, “Fpocket: An open source platform for ligand pocket detection”, BMC Bioinformatics, 2009, 10:168
Peter Schmidtke, Xavier Barril “Understanding and Predicting Druggability. A High-Throughput Method for Detection of Drug Binding Sites”, J. Med. Chem., 2010, 53 (15), pp 5858–5867
instructions to download and install fpocket can be found HERE
Current version of targetDB works with fpocket3
targetDB will not be able to perform pocket search on windows as fpocket is not available on windows platform